

We have developed three GNAT systems for extracting biomedical category and relation facts from entity-centric corpora, targeted at large unstructured corpora in particular. For drugs, our target corpus is DailyMed, published by NLM. For diseases, our target corpus is WikiDisease.
The following ontology diagram describes the categories and relations of interest, as well as which entity-centric corpora contain documents on entities of each category:
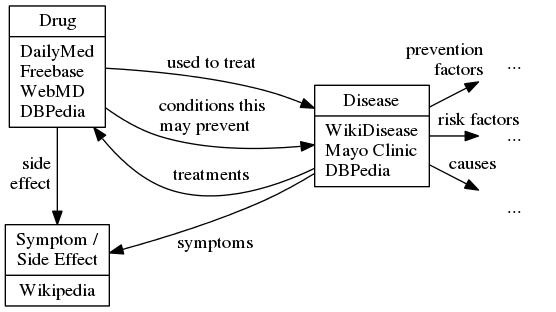
DIEL is restricted to the DailyMed corpus for inference and Freebase for seeds, and is chiefly concerned with classifying entity mentions according to type. It uses coordinate lists as coupling entities to effect label propagation from the seeds to unlabeled DailyMed entities in a semi-supervised fashion. The most confident labels in the result are used to train a classifier over lexical features of the entity mentions (tokens, suffixes/prefixes, bigrams, head verb, verb modifiers, verp path). DIEL outperforms naive distant-supervision and label-propagation baselines for recall over a held-out set of Freebase seeds.
DIEBOLDS makes use of both structured and unstructured corpora, and classifies entity mentions by their relationship to the title entity of the article. The smaller structured corpus (WebMD, Mayo Clinic) is used to bootstrap SSL over the larger unstructured DailyMed corpus. Articles in the structured corpus are already organized into relevant sections such as "Side Effects" "Risk Factors" etc. In addition to coordinate lists, DIEBOLDS uses section titles as coupling entities in the structured corpora, as well as "neighbor" coupling, our term for a high bag-of-words similarity between an NP context in the target corpus and one in the structured corpus. Label propagation, pruning, and a second classifier proceed following the DIEL system. DIEBOLDS outperforms several naive distant-supervision models for recall and precision over a set of human-generated labels.
The DIEJOB system further refines DIEBOLDS. First, relation seeds are restricted by their section in the structured corpus. Second, DIEJOB generates category seeds by label propagation on coordinate lists from the relation seeds, using the range type of the relation and filtering by section in the structured corpus. Third, the label propagation graph for relation mentions includes only coordinate lists and lexical features, combining the features from both phases of DIEL and excluding the document-structure and neighbor coupling nodes from DIEBOLDS. The edges of this graph are annotated with their TFIDF weight, treating mentions as documents and features as words. Label propagation, pruning, and a second classifier proceed in a now-familiar fashion. DIEJOB outperforms several naive distant-supervision models, DIEBOLDS, MultiR, MIML-RE, and Mintz++ on F1 over a set of human-generated labels.